News
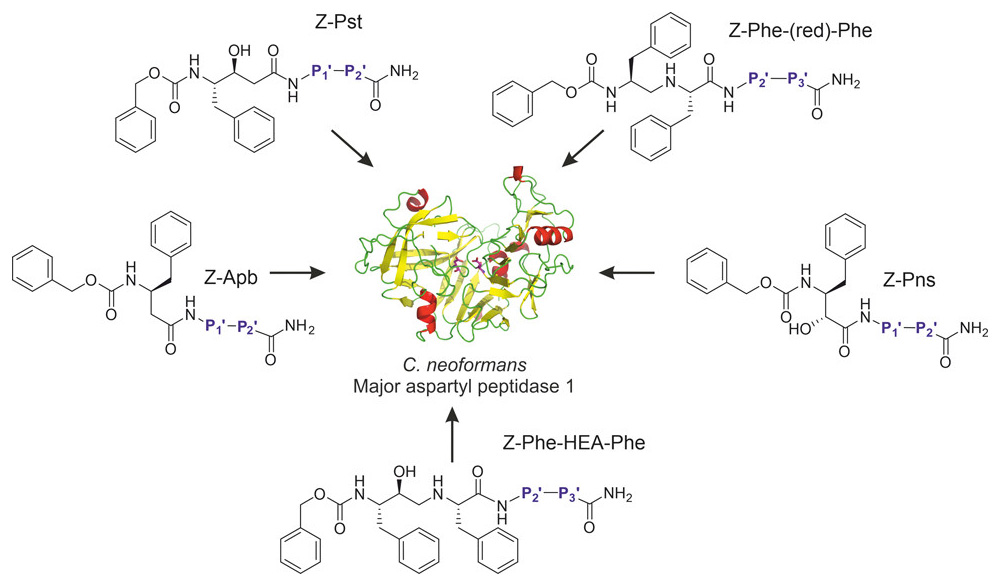
We have a new paper “Re-emerging aspartic protease targets: Examining Cryptococcus neoformans major aspartyl peptidase 1 as a target for antifungal drug discovery“ in the Journal of Medicinal Chemistry.
Congrats to Robin!
Citation: Kryštůfek, R.; Šácha, P.; Starková, J.; Brynda, J.; Hradilek, M.; Tloušt’ová, E.; Grzymska, J.; Rut, W.; Boucher, M. J.; Drąg, M.; Majer, P.; Hájek, M.; Řezáčová, P.; Madhani, H. D.; Craik, C. S.; Konvalinka, J. Re-emerging Aspartic Protease Targets: Examining Cryptococcus neoformans Major Aspartyl Peptidase 1 as a Target for Antifungal Drug Discovery. J. Med. Chem. 2021, 64, 6706-6719. https://doi.org/10.1021/acs.jmedchem.0c02177
Abstract: Cryptococcosis is an invasive infection that accounts for 15% of AIDS-related fatalities. Still, treating cryptococcosis remains a significant challenge due to the poor availability of effective antifungal therapies and emergence of drug resistance. Interestingly, protease inhibitor components of antiretroviral therapy regimens have shown some clinical benefits in these opportunistic infections. We investigated Major aspartyl peptidase 1 (May1), a secreted Cryptococcus neoformans protease, as a possible target for the development of drugs that act against both fungal and retroviral aspartyl proteases. Here, we describe the biochemical characterization of May1, present its high-resolution X-ray structure, and provide its substrate specificity analysis. Through combinatorial screening of 11,520 compounds, we identified a potent inhibitor of May1 and HIV protease. This dual-specificity inhibitor exhibits antifungal activity in yeast culture, low cytotoxicity, and low off-target activity against host proteases and could thus serve as a lead compound for further development of May1 and HIV protease inhibitors.
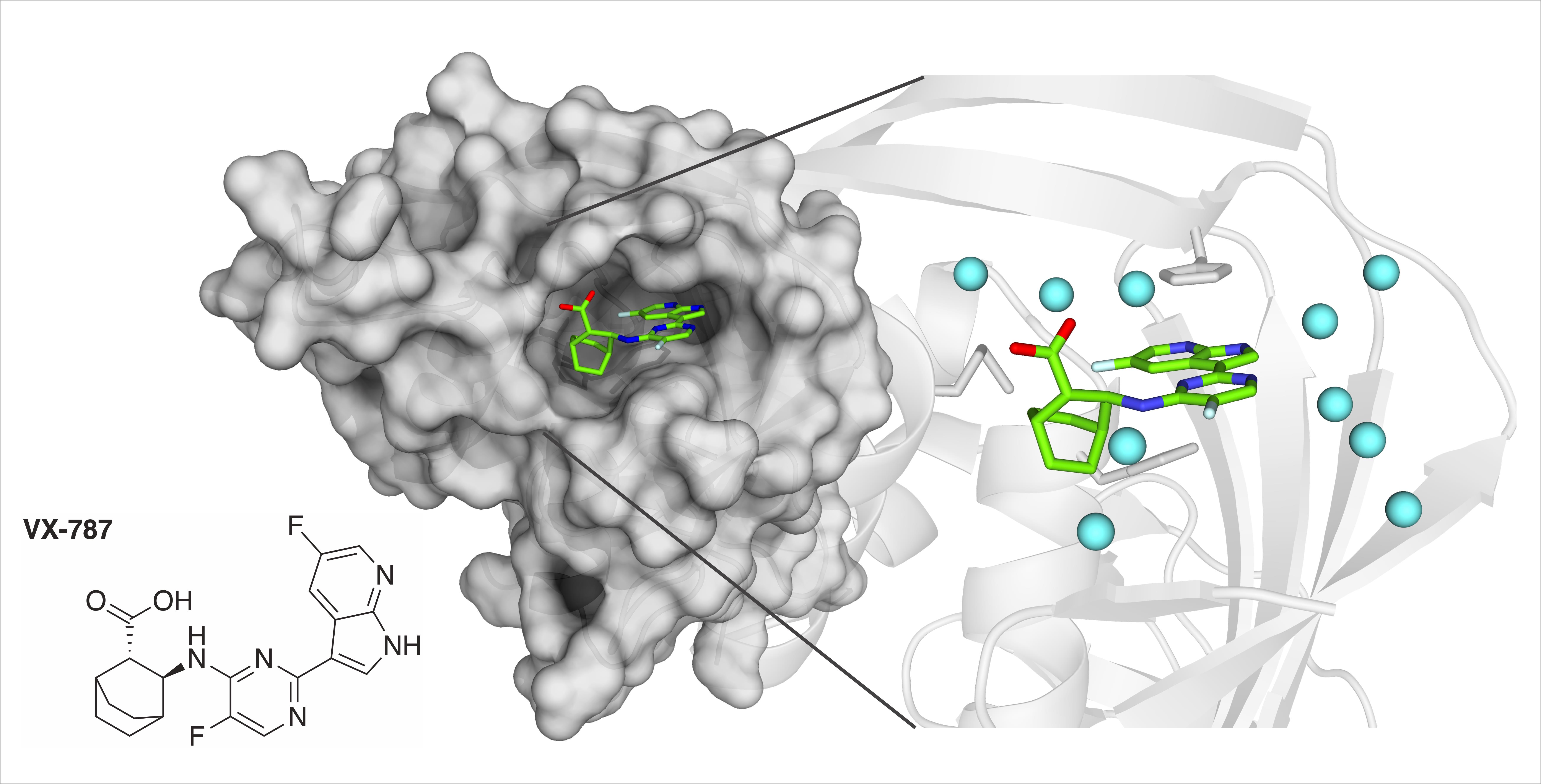
We have a new paper “Thermodynamic Analysis of the Resistance Development to Pimodivir (VX-787), the Clinical Inhibitor of Cap Binding to PB2 Subunit of Influenza A Polymerase“ in the Molecules.
Congrats to Jirka!
Citation: Gregor, J.; Radilová, K.; Brynda, J.; Fanfrlík, J.; Konvalinka, J.; Kožíšek, M. Structural and Thermodynamic Analysis of the Resistance Development to Pimodivir (VX-787), the Clinical Inhibitor of Cap Binding to PB2 Subunit of Influenza A Polymerase. Molecules 2021, 26. https://doi.org/10.3390/molecules26041007
Abstract: Influenza A virus (IAV) encodes a polymerase composed of three subunits: PA, with endonuclease activity, PB1 with polymerase activity and PB2 with host RNA five-prime cap binding site. Their cooperation and stepwise activation include a process called cap-snatching, which is a crucial step in the IAV life cycle. Reproduction of IAV can be blocked by disrupting the interaction between the PB2 domain and the five-prime cap. An inhibitor of this interaction called pimodivir (VX-787) recently entered the third phase of clinical trial; however, several mutations in PB2 that cause resistance to pimodivir were observed. First major mutation, F404Y, causing resistance was identified during preclinical testing, next the mutation M431I was identified in patients during the second phase of clinical trials. The mutation H357N was identified during testing of IAV strains at Centers for Disease Control and Prevention. We set out to provide a structural and thermodynamic analysis of the interactions between cap-binding domain of PB2 wild-type and PB2 variants bearing these mutations and pimodivir. Here we present four crystal structures of PB2-WT, PB2-F404Y, PB2-M431I and PB2-H357N in complex with pimodivir. We have thermodynamically analysed all PB2 variants and proposed the effect of these mutations on thermodynamic parameters of these interactions and pimodivir resistance development. These data will contribute to understanding the effect of these missense mutations to the resistance development and help to design next generation inhibitors.
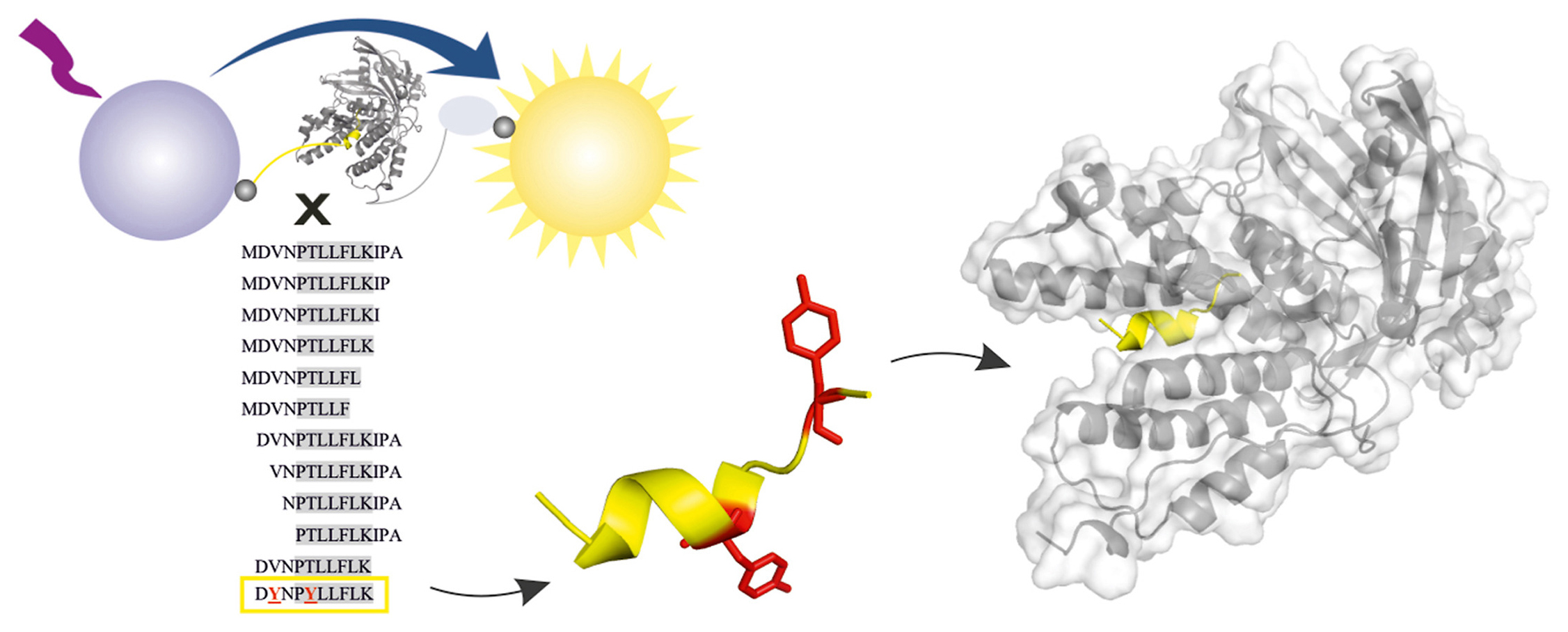
We have a new paper in Antiviral Research. Congrats to Milan, Jakub, and the whole team!
Citation: Hejdánek, J.; Radilová, K.; Pachl, P.; Hodek, J.; Machara, A.; Weber, J.; Řezáčová, P.; Konvalinka, J.; Kožíšek, M. Structural characterization of the interaction between the C-terminal domain of the influenza polymerase PA subunit and an optimized small peptide inhibitor. Antiviral Research 2021, 185, 104971. https://doi.org/10.1016/j.antiviral.2020.104971
Abstract: Influenza viruses can cause severe respiratory infections in humans, leading to nearly half a million deaths worldwide each year. Improved antiviral drugs are needed to address the threat of development of novel pandemic strains. Current therapeutic interventions target three key proteins in the viral life cycle: neuraminidase, the M2 channel and RNA-dependent-RNA polymerase. Protein-protein interactions between influenza polymerase subunits are potential new targets for drug development. Using a newly developed assay based on AlphaScreen technology, we screened a peptide panel for protein-protein interaction inhibitors to identify a minimal PB1 subunit-derived peptide that retains high inhibition potential and can be further modified. Here, we present an X-ray structure of the resulting decapeptide bound to the C-terminal domain of PA polymerase subunit from pandemic isolate A/California/07/2009 H1N1 at 1.6 Å resolution and discuss its implications for the design of specific, potent influenza polymerase inhibitors.
We have a new paper "Unraveling the anti-influenza effect of flavonoids: Experimental validation of luteolin and its congeners as potent influenza endonuclease inhibitors" in the European Journal of Medicinal Chemistry.
Congrats to Katka, Milan, and the whole team!
Citation: Zima, V.; Radilová, K.; Kožíšek, M.; Albiñana, C. B.; Karlukova, E.; Brynda, J.; Fanfrlík, J.; Flieger, M.; Hodek, J.; Weber, J.; Majer, P.; Konvalinka, J.; Machara, A. Unraveling the anti-influenza effect of flavonoids: Experimental validation of luteolin and its congeners as potent influenza endonuclease inhibitors. Eur. J. Med. Chem. 2020, 208, 112754. https://doi.org/10.1016/j.ejmech.2020.112754
Abstract: The biological effects of flavonoids on mammal cells are diverse, ranging from scavenging free radicals and anti-cancer activity to anti-influenza activity. Despite appreciable effort to understand the anti-influenza activity of flavonoids, there is no clear consensus about their precise mode-of-action at a cellular level. Here, we report the development and validation of a screening assay based on AlphaScreen technology and illustrate its application for determination of the inhibitory potency of a large set of polyols against PA N-terminal domain (PA-Nter) of influenza RNA-dependent RNA polymerase featuring endonuclease activity. The most potent inhibitors we identified were luteolin with an IC50 of 72 ± 2 nM and its 8-C-glucoside orientin with an IC50 of 43 ± 2 nM. Submicromolar inhibitors were also evaluated by an in vitro endonuclease activity assay using single-stranded DNA, and the results were in full agreement with data from the competitive AlphaScreen assay. Using X-ray crystallography, we analyzed structures of the PA-Nter in complex with luteolin at 2.0 Å resolution and quambalarine B at 2.5 Å resolution, which clearly revealed the binding pose of these polyols coordinated to two manganese ions in the endonuclease active site. Using two distinct assays along with the structural work, we have presumably identified and characterized the molecular mode-of-action of flavonoids in influenza-infected cells.
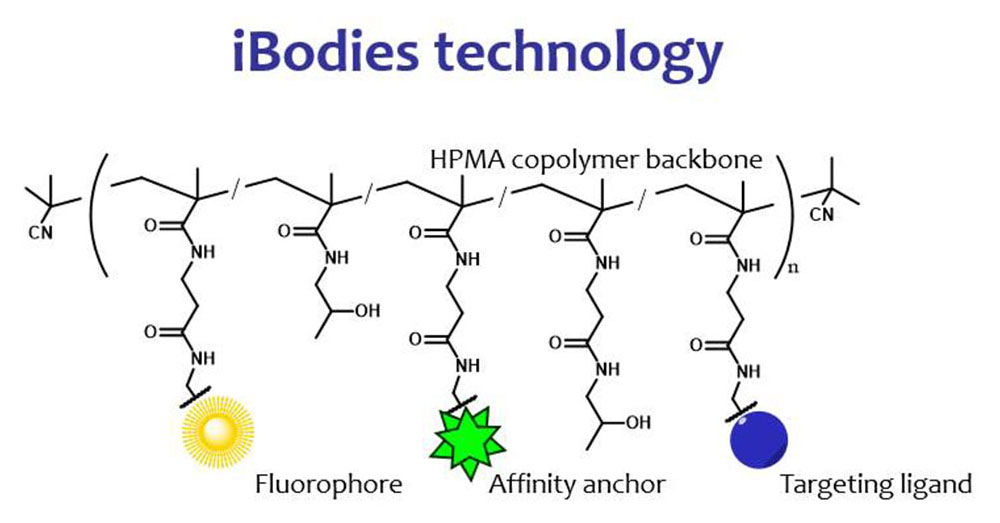
IOCB Prague, the Institute of Macromolecular Chemistry of the Czech Academy of Sciences (IMC), and Charles University have signed a licensing agreement with the company Immunotech, a member of the Beckman Coulter group, for the use of iBodies®, a joint technology for measuring free testosterone in blood.
iBodies were developed by Pavel Šácha, Tomáš Knedlík, and others in our group in collaboration with the team of Vladimír Šubr of the Department of Biomedical Polymers, headed by Tomáš Etrych, at IMC. iBodies comprise a polymer backbone with a small organic molecule bound to it that can bind to the target receptor of tested cells. A component of the complex is a molecule that serves as an anchor for use in various separation techniques and, in some cases, also a fluorescent label facilitating imaging of iBodies with various laboratory methods. All this together gives the technology considerable potential for use in clinical laboratories, either as substitutes for nonsynthetic antibodies that are otherwise not readily available, or as tracers for measurement.
The new iBodies provide an alternative to the radioactive tracer method, thus simplifying the measurement of free testosterone in clinical laboratories.
Working with researchers from the Regional Centre of Advanced Technologies and Materials (RCPTM) at Palacký University and a number of other centers, a team of researchers headed by Pavel Šácha from our group has developed a new method for isolating viral RNA independent of commercially available materials. The researchers optimized the method for laboratory automation in clinical practice, making it possible to isolate nearly a hundred samples at once, and they successfully tested it in collaboration with the Public Health Institute in Ostrava.
We have a new paper in the SLAS Discovery. Congrats to Jan, Václav, and the whole team!
Citation: Tykvart, J.; Navrátil, V.; Kugler, M.; Šácha, P.; Schimer, J.; Hlaváčková, A.; Tenora, L.; Zemanová, J.; Dejmek, M.; Král, V.; Potáček, M.; Majer, P.; Jahn, U.; Brynda, J.; Řezáčová, P.; Konvalinka, J. Identification of Novel Carbonic Anhydrase IX Inhibitors Using High-Throughput Screening of Pooled Compound Libraries by DNA-Linked Inhibitor Antibody Assay (DIANA). SLAS DISCOVERY: Advancing the Science of Drug Discovery 2020, 25, 1026-1037. https://doi.org/10.1177/2472555220918836
Abstract: The DNA-linked inhibitor antibody assay (DIANA) has been recently validated for ultrasensitive enzyme detection and for quantitative evaluation of enzyme inhibitor potency. Here we present its adaptation for high-throughput screening of human carbonic anhydrase IX (CAIX), a promising drug and diagnostic target. We tested DIANA’s performance by screening a unique compound collection of 2816 compounds consisting of lead-like small molecules synthesized at the Institute of Organic Chemistry and Biochemistry (IOCB) Prague (“IOCB library”). Additionally, to test the robustness of the assay and its potential for upscaling, we screened a pooled version of the IOCB library. The results from the pooled screening were in agreement with the initial nonpooled screen with no lost hits and no false positives, which shows DIANA’s potential to screen more than 100,000 compounds per day.
All DIANA screens showed a high signal-to-noise ratio with a Z′ factor of >0.89. The DIANA screen identified 13 compounds with Ki values equal to or better than 10 µM. All retested hits were active also in an orthogonal enzymatic assay showing zero false positives. However, further biophysical validation of identified hits revealed that the inhibition activity of several hits was caused by a single highly potent CAIX inhibitor, being present as a minor impurity. This finding eventually led us to the identification of three novel CAIX inhibitors from the screen. We confirmed the validity of these compounds by elucidating their mode of binding into the CAIX active site by x-ray crystallography.
The team led by Pavel Šácha with František Sedlák, Robin Kryštůfek, and others, in extensive collaboration with scientists from RCPTM, IMIC CAS, UCT Prague, IMG CAS, IEB CAS, BIOCEV, CEITEC, and clinical laboratories at FN Motol, FN Bulovka, National Institute of Public Health, etc. are developing alternative RNA isolation kits for scaling-up COVID-19 diagnostics, as the commercial diagnostic sets are not available at the moment and probably won’t be for foreseeable future.
The researchers are optimizing a method of SARS-CoV-2 RNA isolation based on magnetic beads from Radek Zbořil lab at RCPTM.
The method is now being validated in clinical laboratories and the first results show it is comparable to existing diagnostics kits. The scientists are now preparing sets for thousands of isolations distributed for trial testing to the laboratories that have already run out of them.
The team continues developing the method to allow robotization which is necessary for massive scale-up of testing.
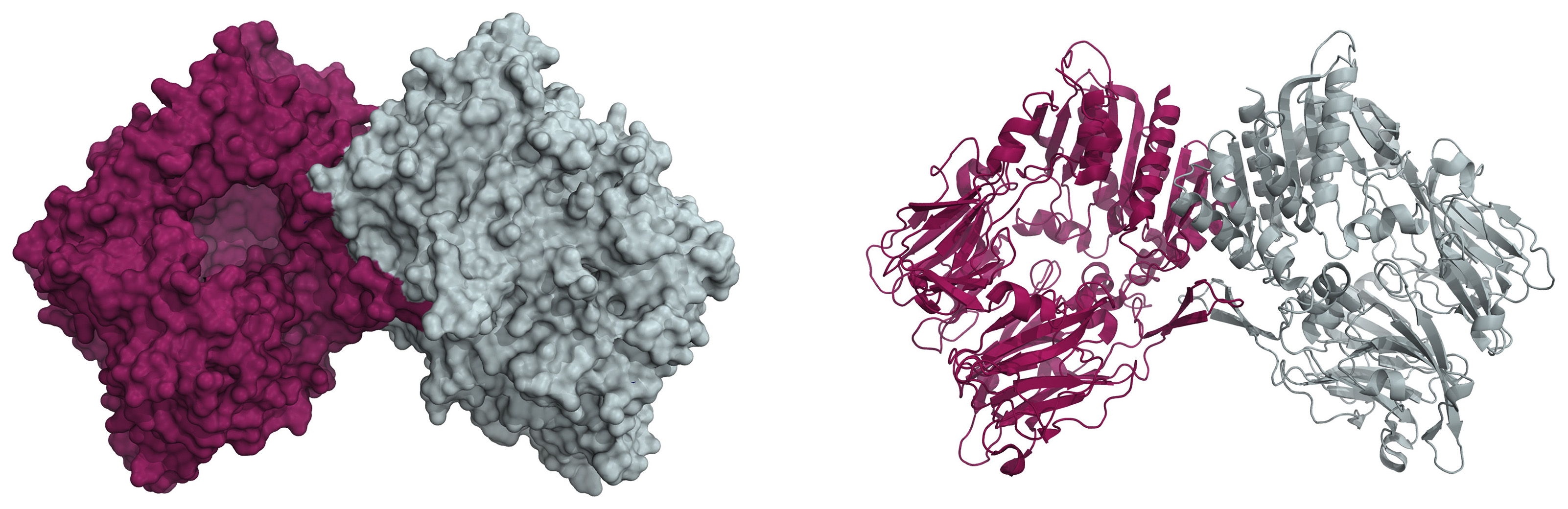
We have a new review in the BBA – Proteins and Proteomics. Congrats to Adéla!
Citation: Šimková, A.; Bušek, P.; Šedo, A.; Konvalinka, J. Molecular recognition of fibroblast activation protein for diagnostic and therapeutic applications. Biochimica et Biophysica Acta (BBA) – Proteins and Proteomics 2020, 1868, 140409. https://doi.org/10.1016/j.bbapap.2020.140409
Abstract: Fibroblast activation protein (FAP) is a non-classical serine protease expressed predominantly in conditions accompanied by tissue remodeling, particularly cancer. Due to its plasma membrane localization, FAP represents a promising molecular target for tumor imaging and treatment. The unique enzymatic activity of FAP facilitates development of diagnostic and therapeutic tools based on molecular recognition of FAP by substrates and small-molecule inhibitors, in addition to conventional antibody-based strategies.
In this review, we provide background on the pathophysiological role of FAP and discuss its potential for diagnostic and therapeutic applications. Furthermore, we present a detailed analysis of the structural patterns crucial for substrate and inhibitor recognition by the FAP active site and determinants of selectivity over the related proteases dipeptidyl peptidase IV and prolyl endopeptidase. We also review published data on targeting of the tumor microenvironment with FAP antibodies, FAP-targeted prodrugs, activity-based probes and small-molecule inhibitors. We describe use of a recently developed, selective FAP inhibitor with low-nanomolar potency in inhibitor-based targeting strategies including synthetic antibody mimetics based on hydrophilic polymers and inhibitor conjugates for PET imaging.
In conclusion, recent advances in understanding of the molecular structure and function of FAP have significantly contributed to the development of several tools with potential for translation into clinical practice.
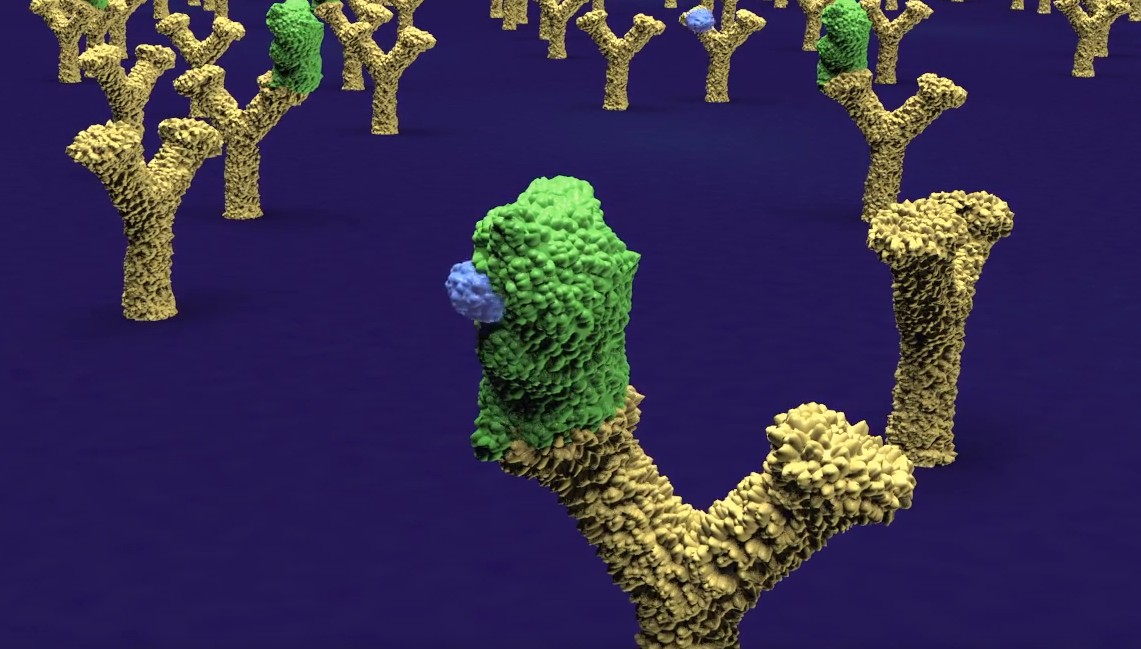
A new company DIANA Biotechnologies has been founded at IOCB Prague to develop a technology known as DIANA, or DNA-linked Inhibitor Antibody Assay, which was developed in our group by Václav Navrátil and colleagues. The unique method allows for ultrasensitive quantification of enzymes and other proteins in a solution, e.g. human blood, urine, or saliva. In practice, it should help doctors diagnose certain diseases (cancer and various infectious diseases, for instance) at an earlier stage than is possible with existing methods. Moreover, the technology has a second important application: It enables screening of large numbers of chemical substances for enzyme inhibitors and the identification of suitable compounds for further development as drug candidates.

